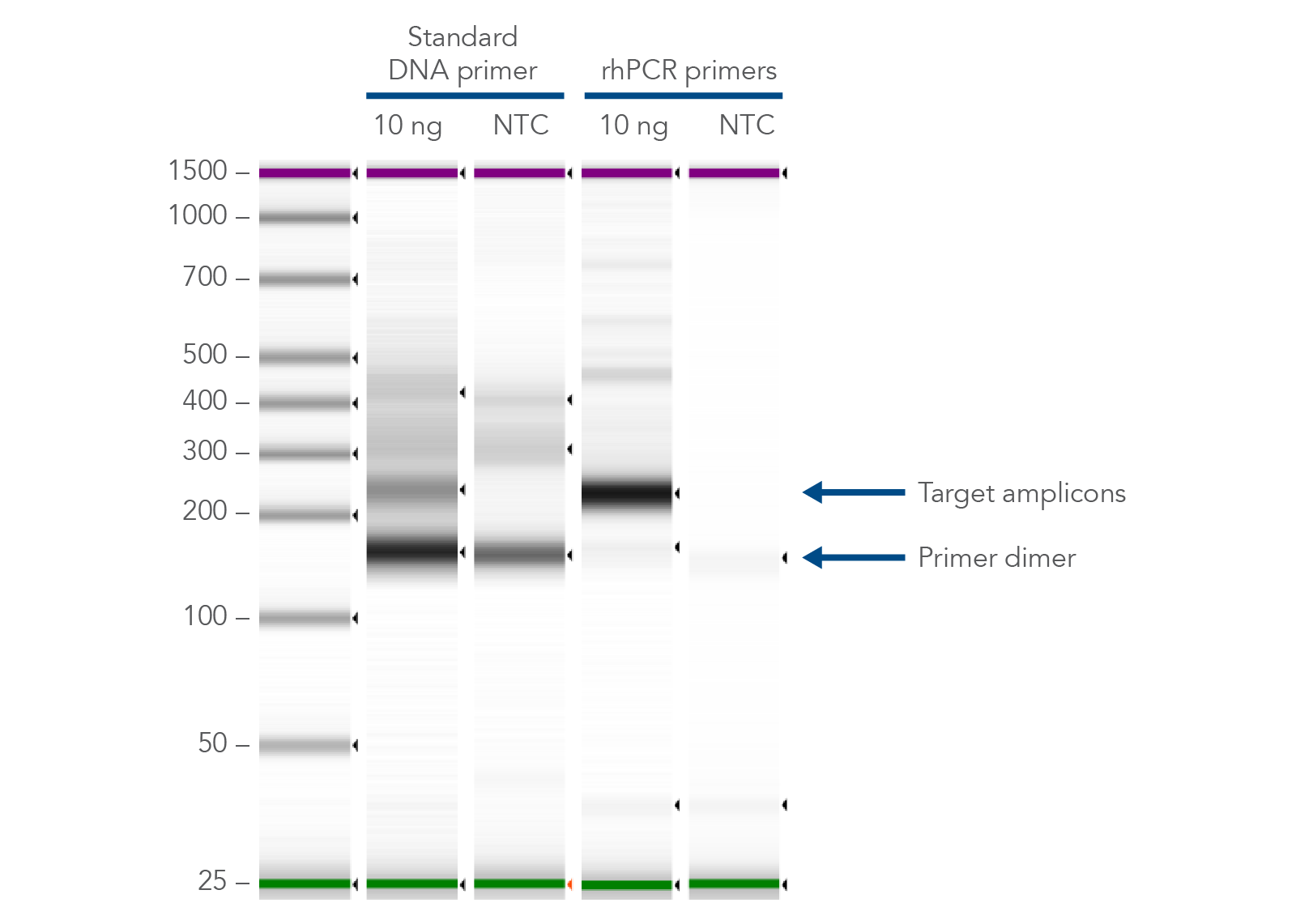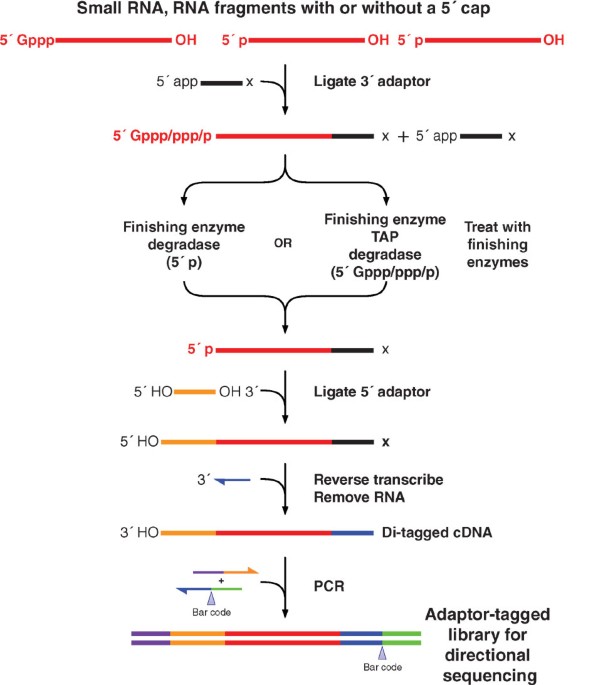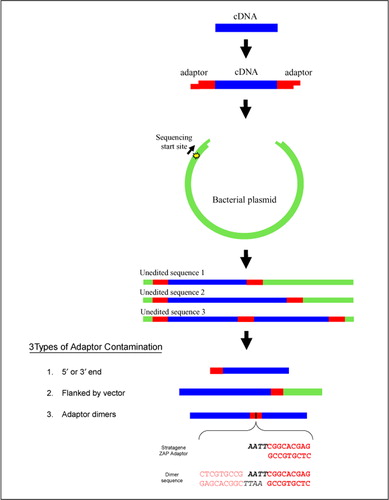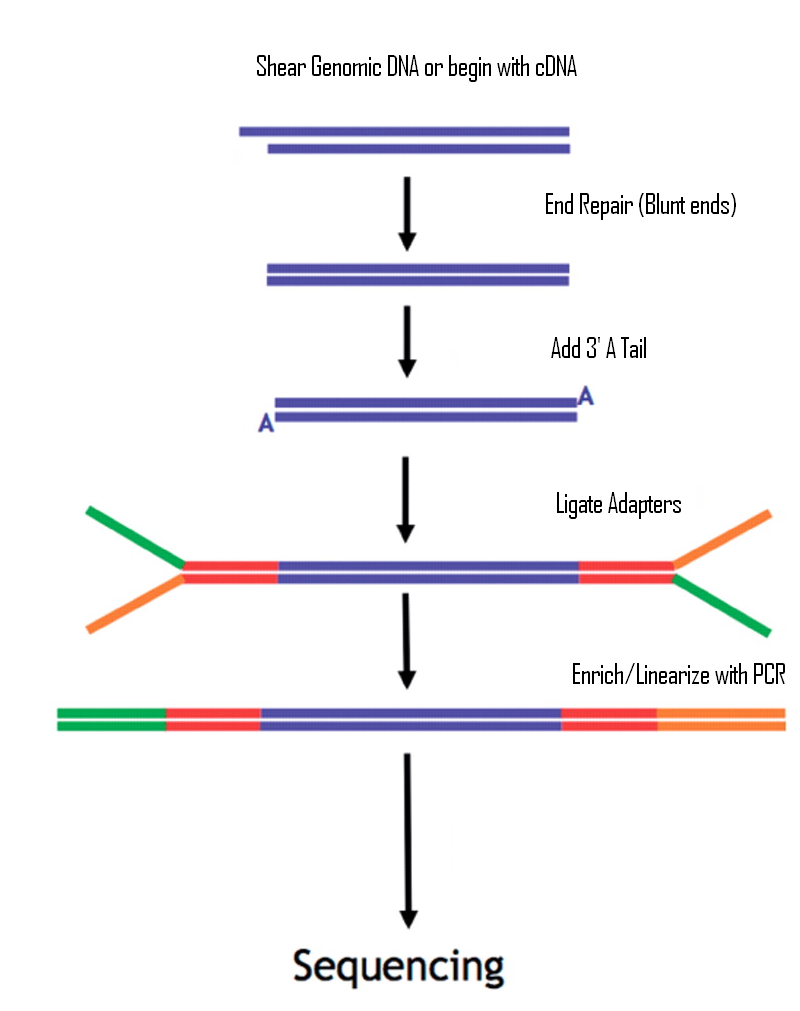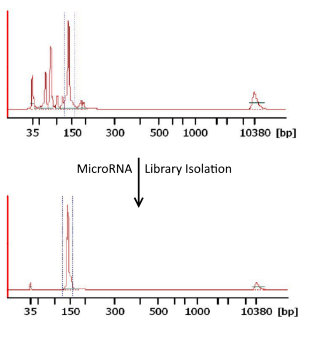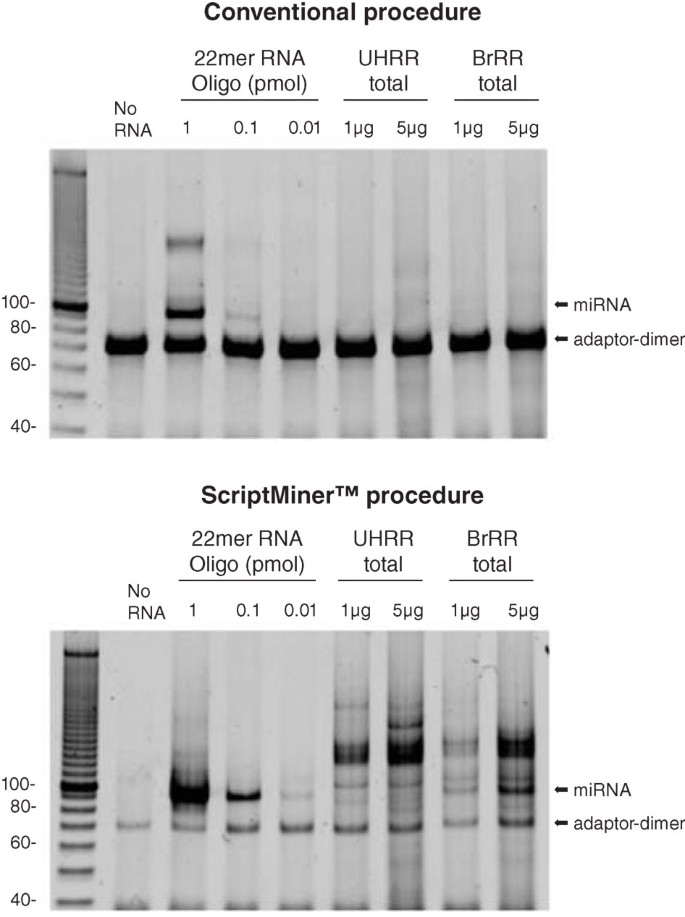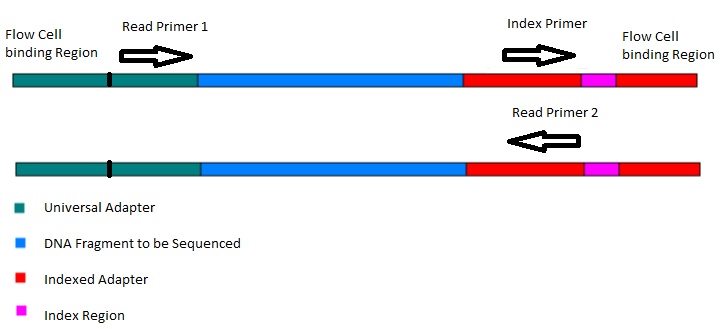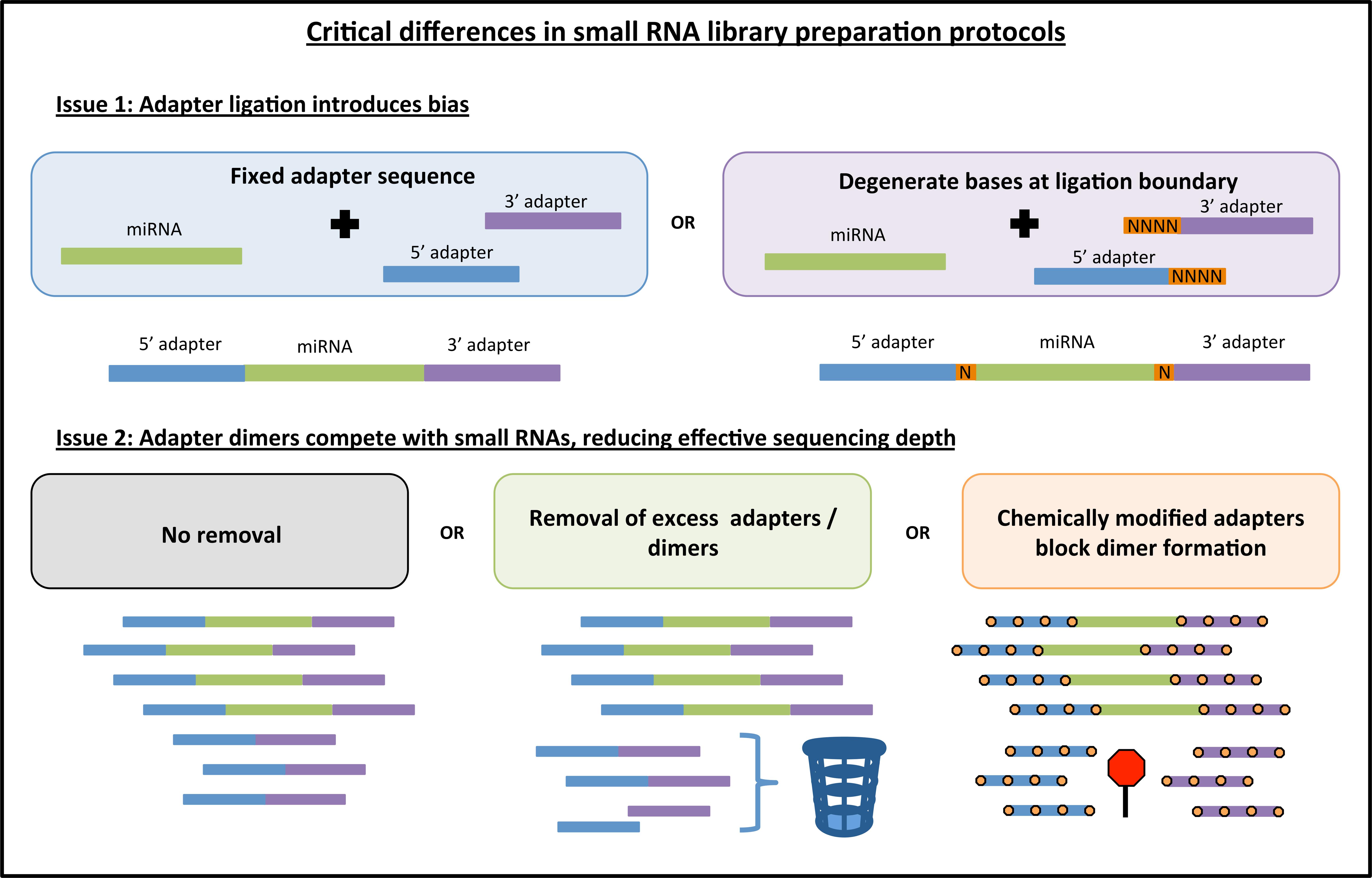
Frontiers | Addressing Bias in Small RNA Library Preparation for Sequencing: A New Protocol Recovers MicroRNAs that Evade Capture by Current Methods
![Methods And Kits For Reducing Adapter-dimer Formation Toloue; Masoud ; et al. [BIOO Scientific Corporation] Methods And Kits For Reducing Adapter-dimer Formation Toloue; Masoud ; et al. [BIOO Scientific Corporation]](https://uspto.report/patent/app/20200190565/US20200190565A1-20200618-D00001.png)
Methods And Kits For Reducing Adapter-dimer Formation Toloue; Masoud ; et al. [BIOO Scientific Corporation]
Small RNA Library Preparation Method for Next-Generation Sequencing Using Chemical Modifications to Prevent Adapter Dimer Formation | PLOS ONE